Antibody Antigen Docking
Antibodies, which bind tightly to specific antigens to block their activity or to mark them for destruction, play essential roles in immune system. Antibodies are a kind of Y-shaped proteins, consisting two pairs of identical polypeptide chains which are light and heavy chains. Based on the properties of structural and sequence variability, it's possible for their variable domains to impart most of the specificity of an antibody for its antigen target. Modern computational approaches can predict antibody-antigen structures, facilitate the design and engineering of antibody-based therapeutics by taking advantages of these variable domains.
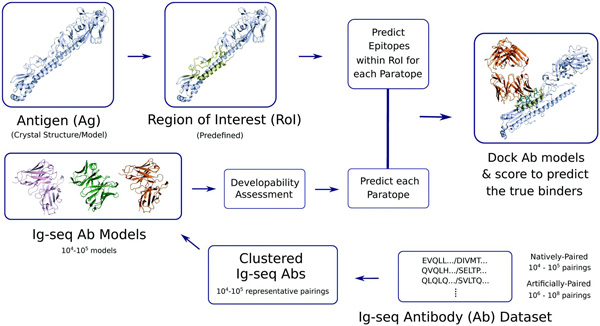 Fig.1 A general procedures of antibody-antigen docking. (Raybould, M.; et al. 2019)
Fig.1 A general procedures of antibody-antigen docking. (Raybould, M.; et al. 2019)Our Services of Antibody Antigen Docking
Docking
A rigid body searching is performed and we develop rational antibody-antigen complex models. Moreover, user-defined restraints which can be obtained from experimental data are used to limit the search space.
Scoring and ranking
The models obtained from the docking step are scored and ranked using a scoring function which is developed specifically for antibody systems.
Optimization and validation
The structures with highest score are clustered and the largest ones are progressed into the process of structural optimization with the application of energy minimization.
Our Abilities of Antibody Antigen Docking
We have employed various docking techniques to complete the searching for all possible binding modes in the translational and rotational space between an antibody (Fv, Fab, or full antibody) and its antigen.
Our antibody teams can design models of conformational changes of the variable loops upon complex formation.
We also provide binding affinities which are experimentally determined and other additional services such as antibody affinity prediction and algorithms design.
At BOC Sciences, we have state-of-the-art computational tools for modeling and refinement of Fv loop regions.
Computational epitope mapping is available to identify the antibody-antigen interactions.
Our docking procedures are compatible with different antibody numbering schemes for immunoglobulin variable domains, such as Chothia, Kabat, IMGT, Aho.
Our experts can conduct model quality assessment by multiple criteria.
We have rich experiences in performing various antibody-antigen cases as well as other classes of protein complexes.
Reference
Raybould, M.; et al. Antibody–antigen complex modelling in the era of immunoglobulin repertoire sequencing. Molecular Systems Design & Engineering. 2019, 4(4): 679-688.
※ It should be noted that our service is only used for research.

One-stop
Drug Discovery Services
- Experienced and qualified scientists functioning as project managers or study director
- Independent quality unit assuring regulatory compliance
- Methods validated per ICH GLP/GMP guidelines
- Rigorous sample tracking and handling procedures to prevent mistakes
- Controlled laboratory environment to prevent a whole new level of success
Online Inquiry

