Structure-Based Virtual Screening
SBVS approaches are applied as in silico experiments for the rapid and accurate identification of promising hits. When searching for new hits in SBVS, 3D structure of the target is utilized and usually can be obtained from biophysical methods (X-ray crystallography and NMR spectroscopy) or modeling methods. Scientists then use the target structure to docking a large amount of chemical molecules and identify the most potential hit candidates.
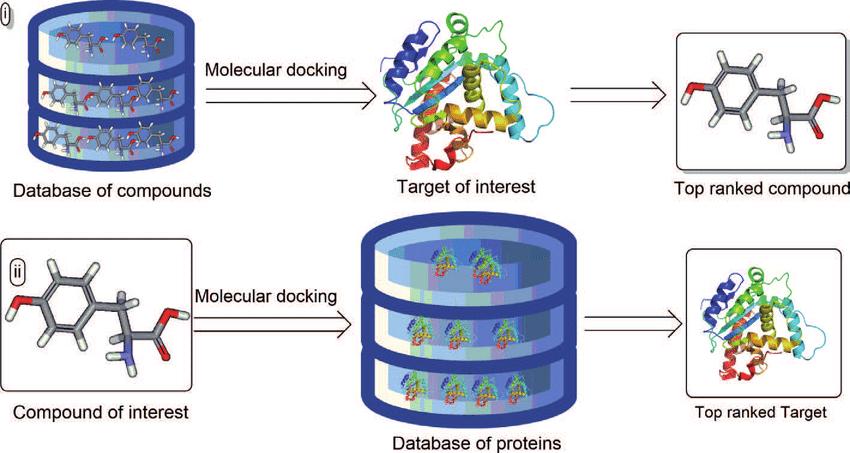 Fig.1 Two procedures of structure-based virtual screening. (Badria, F. A.; Elgazar, A. A. 2020)
Fig.1 Two procedures of structure-based virtual screening. (Badria, F. A.; Elgazar, A. A. 2020)Advantages of SBVS
Time-saved and cost-effective.
Examination can be conducted computationally without the experimental synthesis steps.
Various computational programs or tools can be applied and provide assist in SBVS.
SBVS Procedures at BOC Sciences
Target selection
We conduct the searching of determined structures in various public chemical databases and we can also obtain the target protein by performing homology modeling.
Libraries screening
At BOC Sciences, multiple public resources and commercial chemical collections are available for us to carry out large-scale SBVS. And we also have designed high-quality virtual libraries which contain different chemical molecules.
Docking and analysis
Both molecular docking and molecular dynamics simulations can be used to screen out a series of hits and corresponding binding modes.
Our Capabilities of SBVS Services
Homology modeling
As a most widely-used computational simulation method, homology modeling approach is known as the most accurate, reliable and well-established approach of computer modeling. At BOC Sciences, we apply this method to predict protein structures, supporting the subsequent docking procedure and other experimental work.
Molecular docking
Scientists use molecular docking to establish models at the atomic level, allowing for studies of the interactions between proteins and other molecules as well as mechanism of such interaction. Our bioinformatic experts help to predict the 3D structure of these interacting partners with the application of computer-aided docking technology.
Molecular dynamics (MD) simulation
This method studies the protein structure and dynamics by simulating the dynamics of a collection of atoms, molecules, particles and so on. The structural, dynamical, thermodynamic and statistical properties of protein-ligand complexes can be calculated in this simulation. Aiming to help our customers better understand the molecular mechanisms and interactions on the atomic scale, our groups study ligand-protein or protein-protein complexes and other processes. Combined with advanced computing tools and MD simulation algorithms, we are capable of investigating the dynamics of multiple target proteins, including soluble proteins, membrane proteins, and large molecular machines.
Our Advantages of SBVS Services
Our groups improve the success rate of the in vitro experiments and enhance the hit rate by decreasing the number of compounds which are estimated for their activity experimentally using SBVS methods.
At BOC Sciences, our SBVS platforms have equipped with state-of-the-art computing equipment and advanced software tools to support each stage of the SBVS procedure.
In addition to commercial libraries and our own collections, we can also offer customized libraries according to the demand of your project.
Our experts have rich experience in the application of SBVS for novel drug development and targeted treatment planning.
Reference
Badria, F. A.; Elgazar, A. A. Drug Repurposing in Dermatology: Molecular Biology and Omics Approach. 2020.
※ It should be noted that our service is only used for research.

One-stop
Drug Discovery Services
- Experienced and qualified scientists functioning as project managers or study director
- Independent quality unit assuring regulatory compliance
- Methods validated per ICH GLP/GMP guidelines
- Rigorous sample tracking and handling procedures to prevent mistakes
- Controlled laboratory environment to prevent a whole new level of success
Online Inquiry

