Protein Lipid Docking
A great numbers of drug targets are embedded within the phospholipid bilayer of cellular membranes, including G protein-coupled receptors, ion channels, transporters and membrane-bound enzymes. Structural studies have shown that various small-molecule drugs interact with these targets at binding sites at the protein-phospholipid interface. The membrane lipids activate the bioactivity of proteins by stabilizing the structure and promoting rearrangement, assembly, dissociation, or conformational changes and these membrane proteins can therefore regulate many important cellular events. Nowadays, computational docking method has proved a rapid and reliable tool to study the ligand-lipid interactions at the atomic level in small-molecule drug action and provide a accurate prediction for new drug design.
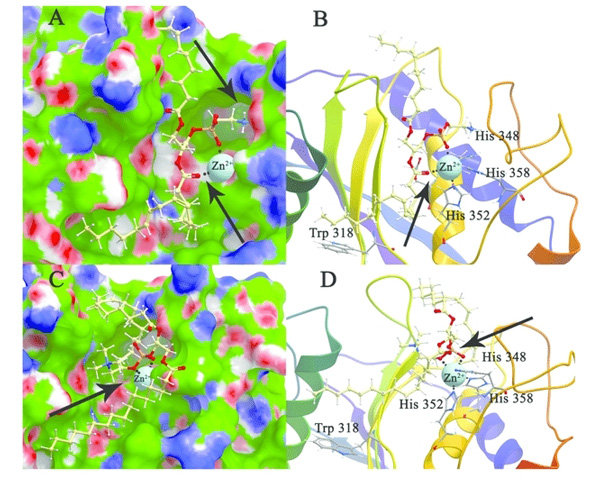 Fig.1 Modeling and docking of protein-lipid interactions. (Zakharzhevskaya, N.; et al. 2017)
Fig.1 Modeling and docking of protein-lipid interactions. (Zakharzhevskaya, N.; et al. 2017)Our Services of Protein Lipid Docking
Docking
We deliver the most rational protein-lipid complex combing the sequence and structure information.
Scoring
Our teams have designed and applied the scoring function to predict the lipid distortion and protein conformational changes associated with the binding event.
The searching space can be limited using experiment-derived restraints.
Optimization
We select the structures with best values of binding energies and the largest clusters are progressed into the following-up structural optimization through energy minimization.
Our Advantages of Protein Lipid Docking
We use multiple computer algorithms to search for amino acid sequence and generate lipid binding sites of a protein.
Our capabilities are also included the three-dimensional structure modeling using docking approaches, which is necessary to identify the region of the protein where the lipid associates or binds.
We can monitor and assess various binding mechanisms in protein-lipid interactions including covalent, hydrophobic, electrostatics, hydrogen bonds, van der Waals interactions and ionic bridges between the protein and lipid ligand.
At BOC Sciences, multiple appropriate lipid types and compositions are available for your docking experiments.
Our scientists are also capable of simulating the effect on membrane distortion and protein structural changes.
We support the integration with downstream molecular dynamic simulations in which the final docked complexes can be further tested.
We help to study the membrane access mechanism and offer important information such as potency data, structure-activity relationships, pharmacokinetics and physicochemical properties for drugs that target these sites.
Reference
Zakharzhevskaya, N.; et al. Interaction of Bacteroides fragilis Toxin with Outer Membrane Vesicles Reveals New Mechanism of Its Secretion and Delivery. frontiers in Cellular and Infection Microbiology. 2017, 7.
※ It should be noted that our service is only used for research.

One-stop
Drug Discovery Services
- Experienced and qualified scientists functioning as project managers or study director
- Independent quality unit assuring regulatory compliance
- Methods validated per ICH GLP/GMP guidelines
- Rigorous sample tracking and handling procedures to prevent mistakes
- Controlled laboratory environment to prevent a whole new level of success
Online Inquiry

