Protein Protein Docking
Protein-protein interactions are responsible for a number of key physiological processes in the living cells and underlie the pathomechanism of many diseases. Protein-protein docking interactions between two protein molecules therefore play essential roles in various kinds of cellular processes, such as metabolic control, signal transduction, gene regulation, and cell communication. This powerful tool helps to dig the detailed structural information of a protein-protein interaction and studies the mechanism of such interaction. It therefore can be applied in the screening of pilot drugs and rational optimization of drug molecular structures by modeling the protein-protein complexes.
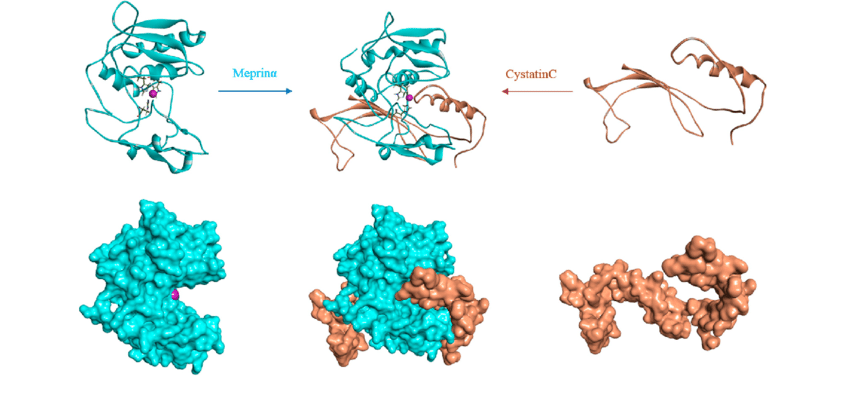 Fig.1 The protein-protein docking model of the meprinα-cystatinC complex
Fig.1 The protein-protein docking model of the meprinα-cystatinC complex(Chaudhuri, A.; et al. 2019)
Our Services of Protein Protein Docking
Docking
We use various algorithm technique to conduct the dockings and generate a set of reliable configurations.
We search for all possible binding modes in the translational and rotational space of the protein-protein complexes,
Calculation and scoring
Energy calculation is applied in the optimization of the binding configurations.
We then conduct energy scoring function for the scoring and ranking procedure.
Optimization and validation
The final optimal conformation is obtained in this step.
Our Capabilities of Protein Protein Docking Services
We are capable of designing and performing Fast Fourier Transform (FFT) based docking algorithm.
Multiple scoring function are available including shape complementarity, electrostatics and desolvation.
We have abilities in performing clustering to find conformations with relative low-energy.
Our docking methods also support the modification of amino acids in proteins for the optimization process.
The final complex structure can be further optimized by energy minimization operation.
Reference
Chaudhuri, A.; et al. Exploring protein-protein intermolecular recognition between meprin-α and endogenous protease regulator cystatinc coupled with pharmacophore elucidation. Journal of Biomolecular Structure & Dynamics. 2019, 37(2): 440-453.
※ It should be noted that our service is only used for research.

One-stop
Drug Discovery Services
- Experienced and qualified scientists functioning as project managers or study director
- Independent quality unit assuring regulatory compliance
- Methods validated per ICH GLP/GMP guidelines
- Rigorous sample tracking and handling procedures to prevent mistakes
- Controlled laboratory environment to prevent a whole new level of success
Online Inquiry

